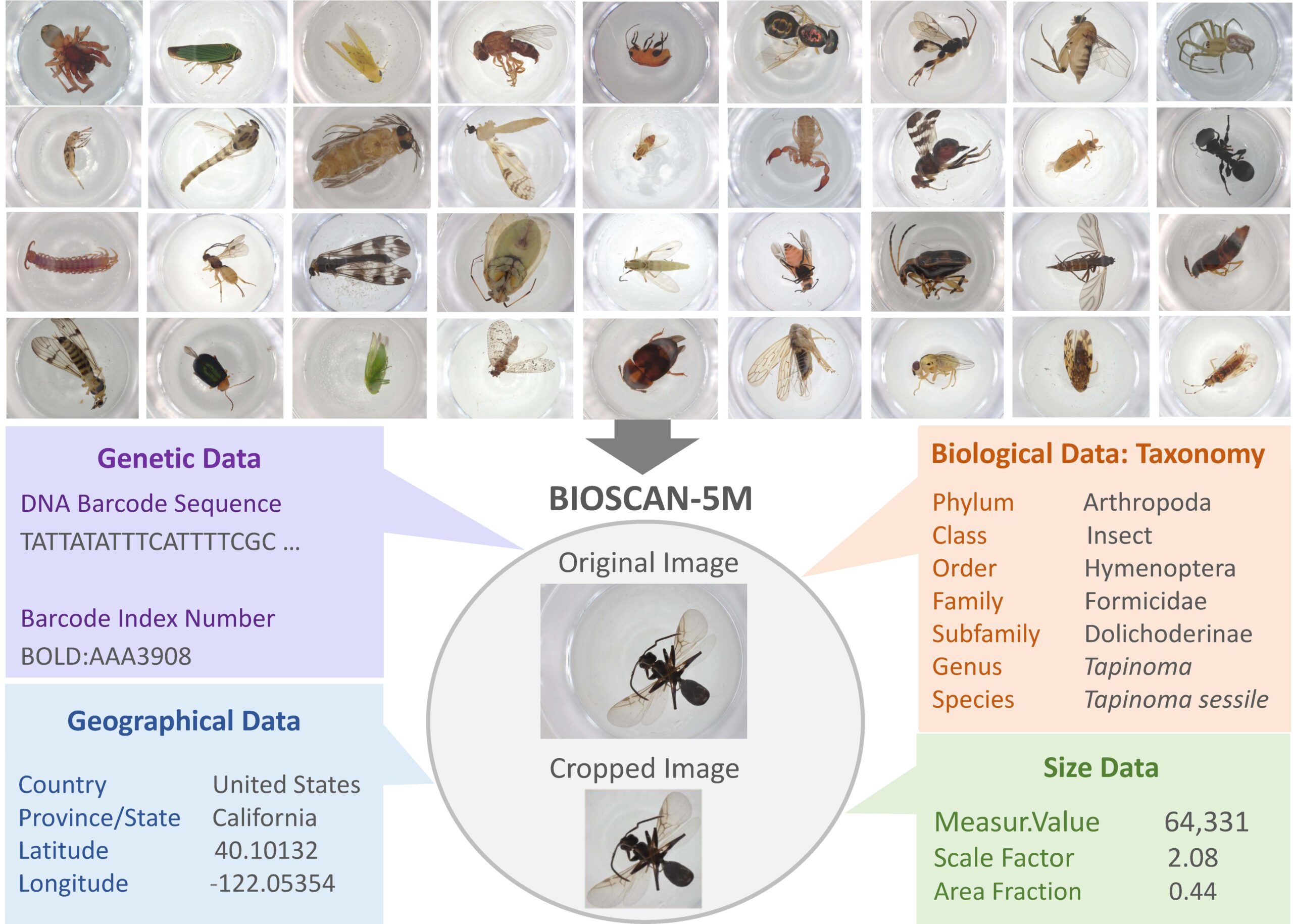
BIOSCAN-5M Insect Dataset
Cataloging insect biodiversity with a new large dataset of hand-labelled insect images
Towards a Taxonomy Machine – A Training Set of 5.6 Million Arthropod Images
This dataset comprises of 5,675,731 images of mostly terrestrial arthropod specimens. The dataset contains images for specimens sampled from 1,698 sites in 48 countries. The image size is 2880 x 2160 pixels before cropping. This translates into an average size of 17.9MB for a tif-file and 1.88 MB for a jpg-file.
The figure shows an array of 95 images taken with a using a Keyence VHX-7000 Digital Microscope system, the empty space at the bottom right corresponds to an empty control well in the source microplate.


BIOSCAN-5M: A Multimodal Dataset for Insect Biodiversity
We propose three benchmark experiments to demonstrate the impact of the multi-modal data types on the classification and clustering accuracy.
First, we pretrain a masked language model on the DNA barcode sequences of the BIOSCAN-5M dataset and demonstrate the impact of using this large reference library on species- and genus-level classification performance.
Second, we propose a zero-shot transfer learning task applied to images and DNA barcodes to cluster feature embeddings obtained from self-supervised learning, to investigate whether meaningful clusters can be derived from these representation embeddings.
Third, we benchmark multi-modality by performing contrastive learning on DNA barcodes, image data, and taxonomic information. This yields a general shared embedding space enabling taxonomic classification using multiple types of information and modalities.

The dataset image packages and metadata are publicly available at:
https://drive.google.com/drive/u/0/folders/1Jc57eKkeiYrnUBc9WlIp-ZS_L1bVlT-0

