Overview
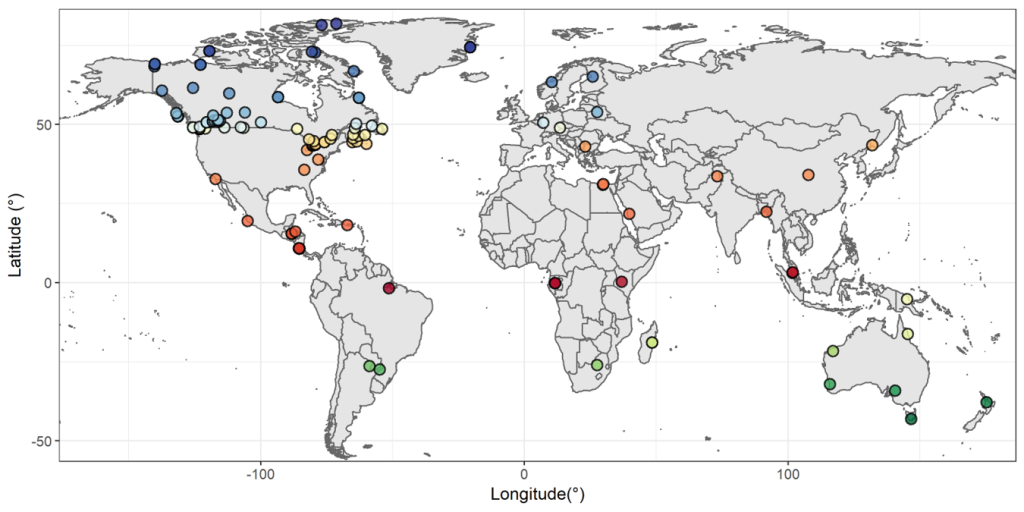
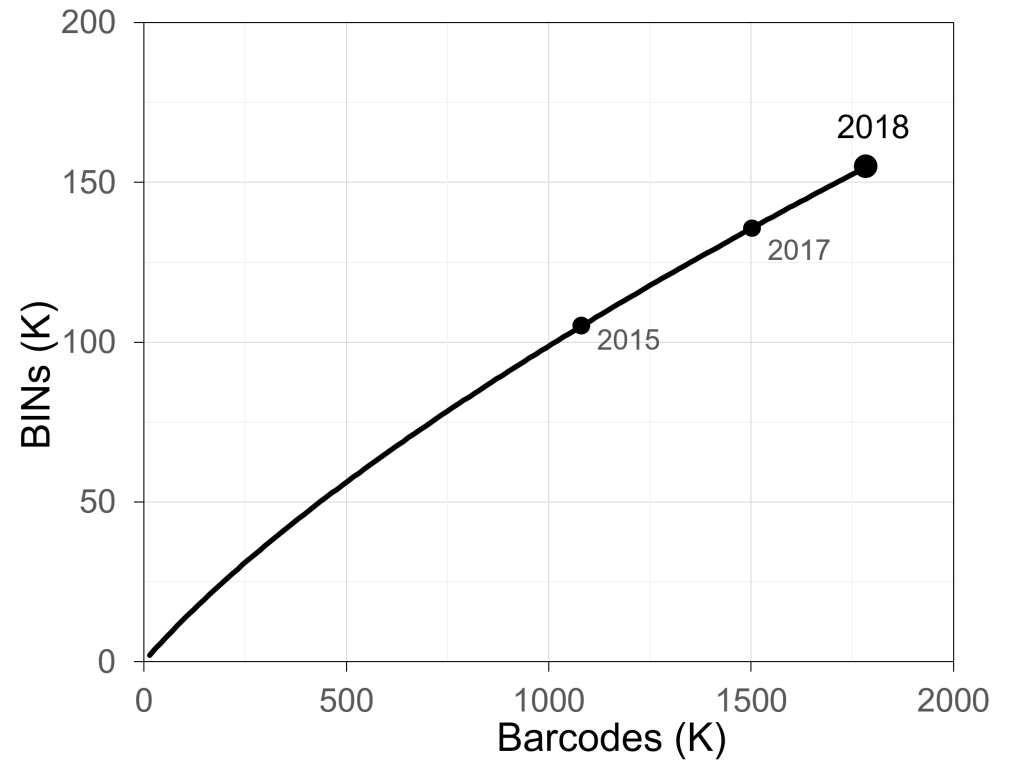
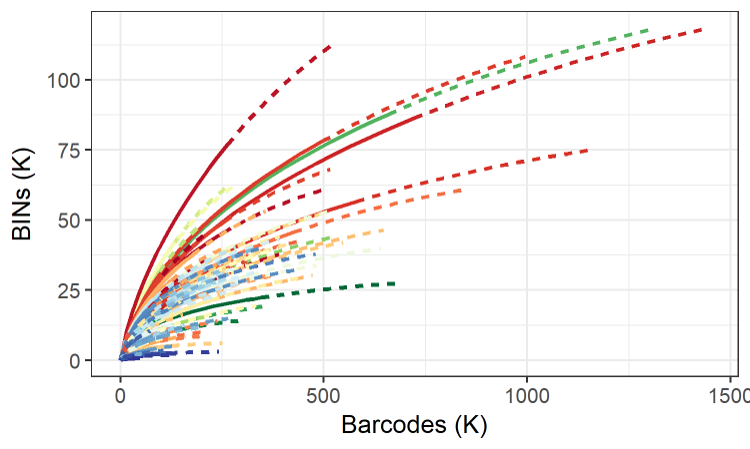
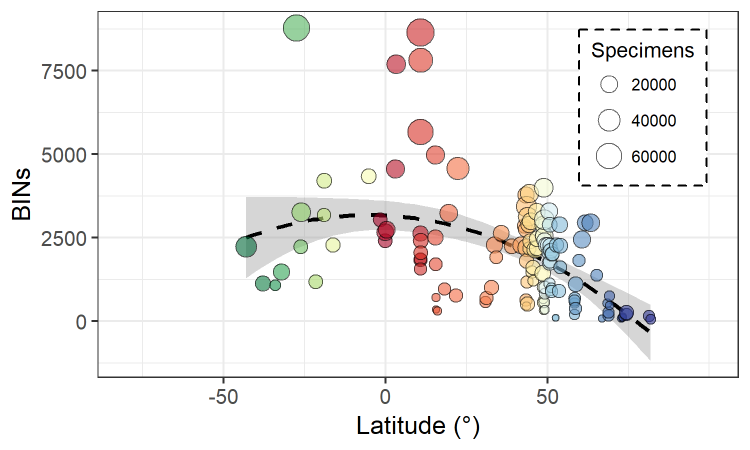
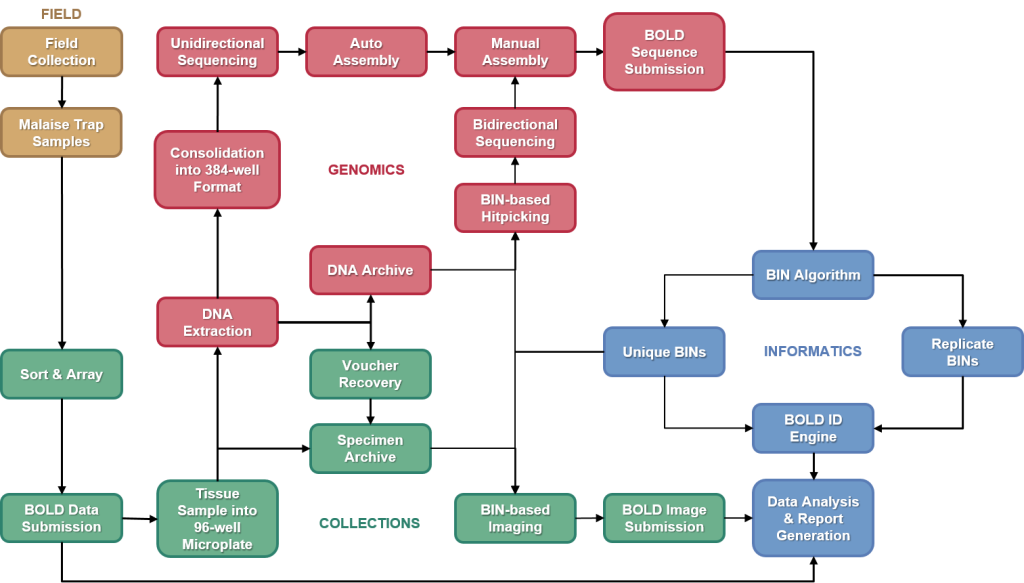
The Global Malaise Trap Program (GMP) was an international collaboration between the CBG and multiple contributors from around the world. Results from the Canadian National Parks (CNP) Malaise Program were incorporated into the larger GMP dataset leading to a total of 158 sampling sites across 33 countries. The program represented the first step toward the acquisition of detailed temporal and spatial information on terrestrial arthropod communities across the globe. Over 2M specimens were sequenced through the GMP, representing over 153K species.
Past efforts to include arthropods in terrestrial assessments faced two barriers: ineffective sampling due to habitat complexity, and unreliable tools for species identification. The second barrier was circumvented by DNA barcoding, a method that utilizes sequence variation in a standardized gene fragment to rapidly sort and objectively differentiate species, while the first barrier was surmounted by the use of Malaise traps as specimen collection tools. Malaise traps are tent-like structures that are effective at capturing insects from various groups and are easily deployed and cost-effective. Used in combination with DNA barcoding, this approach made it possible to carry out large-scale sampling programs and enabled a time- and cost-efficient approach for biodiversity assessments around the world.