Sampling Resources
Experts at the Centre for Biodiversity Genomics at the University of Guelph demonstrate how to prepare museum specimens for DNA Barcoding.
This video makes use of the documents and protocols available for download below.
Museum Sampling Workflow
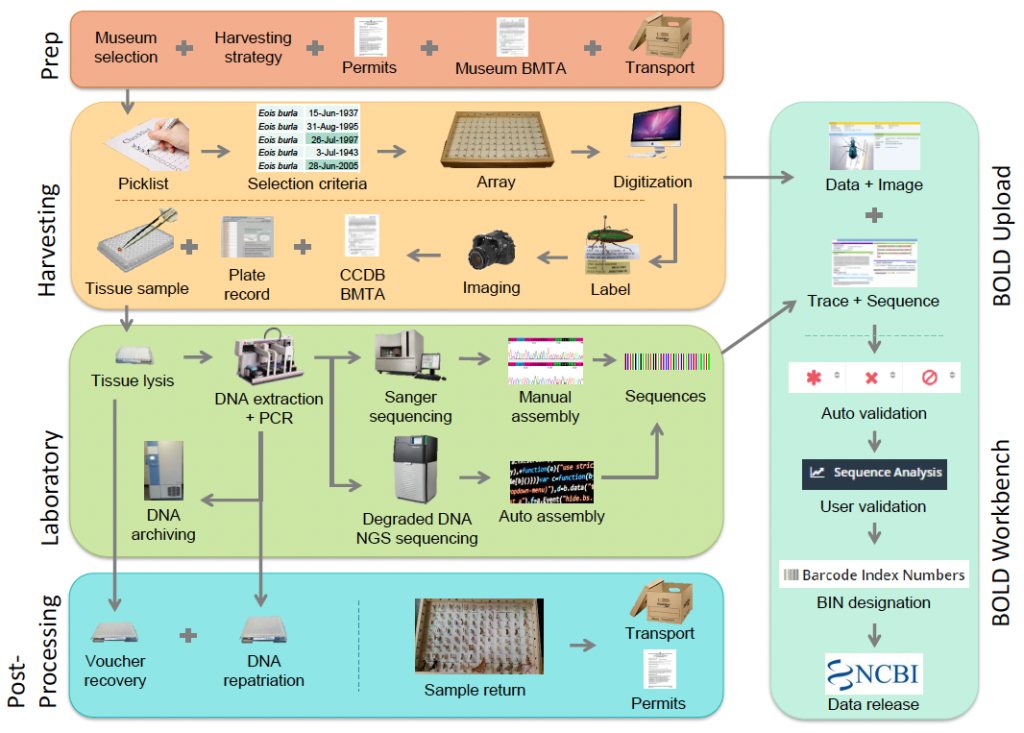
Files provided
Museum Templates
- Museum Label Template (XLS): Create place holders labels for specimen removal from a museum collection, as well as DNA barcode labels to be added to specimens after sampling.
- Museum Harvesting Tracking Sheet (XLS): Spreadsheet to enter auxiliary information (e.g cabinet and taxonomic info) while pulling specimens directly from the Collection.
CCDB and BOLD Templates
- BOLD SpecimenData v3 Submission (XLS): Spreadsheet used to upload specimen data to BOLD.
- BOLD ImageData Submission (XLS): Spreadsheet used to upload image data to BOLD.
- CCDB-00000 Record (XLS): Used to submit plates to CCDB and can be used to print array maps.
- Understanding Sample Submission and Processing at the CCDB.
Documentation
- iBOL-BIO BMTA and Data Policy Agreement (DOC): iBOL Biological Material Transfer and Data Policy Agreement.
- CBG Loan Conditions (PDF): Our loan conditions if you are interested in borrowing specimens.
Shipping
- Protocol for Shipping Ethanol (PDF): International Air Transport Association (IATA) requirements for shipping ethanol.
- NZNHN Shipping Dried Herbarium Specimens (PDF): New Zealand National Herbarium Network – Standard for shipping of dried herbarium specimens.
Additional Resources
iBOL 2017 Workshop Presentation
- Introduction – Natural History Collection Harvesting Strategies & Logistics (PDF)
- Sample Harvesting in Natural History Collections (PDF)
- Specimen Digitization (PDF)
- Pre-lab Processing (PDF)
- Standard Protocols for Natural History Collection Barcoding (PDF)
- Data Analysis and Archiving (PDF)
- New Developments in the lab (PDF)
Other Presentations
- Building a Reference Library: Sourcing Specimens Natural History Collections (PDF): iBOL II talk, Scientific Steering Committee, October 13 & 14, 2018 – deWaard et al.
Lab Resources
http://ccdb.ca/resources/BOLD Resources
http://boldsystems.org/index.php/ResourcesPublications
- Borisenko, A. V., J. E. Sones, and P. D. N. Hebert. 2009. The front-end logistics of DNA barcoding: challenges and prospects. Molecular Ecology Resources 9:27-34. http://onlinelibrary.wiley.com/doi/10.1111/j.1755-0998.2009.02629.x/abstract
- Hausmann, A., S. E. Miller, J. D. Holloway, J. R. deWaard, D. Pollock, S. W. J. Prosser and P. D.N. Hebert. 2016. Calibrating the taxonomy of a megadiverse insect family: 3000 DNA barcodes from geometrid type specimens (Lepidoptera, Geometridae). Genome, 2016, 59(9): 671-684. https://doi.org/10.1139/gen-2015-0197
- Hebert, P. D. N., J. R. deWaard, E. V. Zakharov, S. W. J. Prosser, J. E. Sones, J. T. A. McKeown, B. Mantle, and J. La Salle. 2013. A DNA ‘Barcode Blitz’: Rapid Digitization and Sequencing of a Natural History Collection. Plos One 8:14. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0068535
- Hebert, P. D. N., T. W. A. Braukmann, S. W. J. Prosser, S. Ratnasingham, J. R. deWaard, N. V. Ivanova, D. H. Janzen, W. Hallwachs, S. Naik, J. E. Sones and E. V. Zakharov. 2017. A Sequel to Sanger: Amplicon Sequencing That Scales. BMC Genomics 2018 19:219. https://doi.org/10.1186/s12864-018-4611-3
- Hollingsworth, P. M., D. Li, M. van der Bank and A. D. Twyford. 2016. Telling plant species apart with DNA: from barcodes to genome. Phil. Trans. R. Soc. B 371:20150338. http://dx.doi.org/10.1098/rstb.2015.0338
- Kuzmina, M. L., T. W. A. Braukmann, A. J. Fazekas, S. W. Graham, S. L. Dewaard, A. Rodrigues, B. A. Bennett, T. A. Dickinson, J. M. Saarela, P. M. Catling, S. G. Newmaster, D. M. Percy, E. Fenneman, A. Lauron-Moreau, B. Ford, L. Gillespie, R. Subramanyam, J. Whitton, L. Jennings, D. Metsger, C. P. Warne, A. Brown, E. Sears, J. R. Dewaard, E. V. Zakharov and P. D. N. Hebert. 2017. Using Herbarium-Derived DNAs to Assemble a Large-Scale DNA Barcode Library for the Vascular Plants of Canada. Applications in Plant Sciences, 5(12):1700079. https://doi.org/10.3732/apps.1700079
- Porco, D., R. Rougerie, L. Deharveng, and P. Hebert. 2010. Coupling non-destructive DNA extraction and voucher retrieval for small soft-bodied Arthropods in a high-throughput context: the example of Collembola. Molecular Ecology Resources 10:942-945. http://onlinelibrary.wiley.com/wol1/doi/10.1111/j.1755-0998.2010.2839.x/abstract
- Prosser, S. W. J., J. R. deWaard, S. E. Miller, and P. D. N. Hebert. 2016. DNA barcodes from century-old type specimens using next-generation sequencing. Molecular Ecology Resources 16:487-497. http://onlinelibrary.wiley.com/wol1/doi/10.1111/1755-0998.12474/abstract

